Overview of genetic variation in the Morocco genome
Genotyping and variant invocation of the 109 Moroccan genomes result in an initial VCF file containing 28,262,306 variants containing 24,356,267 SNVs and 4,181,156 indels, with 2,161,454 multiallenes. After applying the GATK VQSR filter, the number of variants decreased to 24,958,854, including 21,760,118 SNVs and 3,400,454 indels, and the multiallensite reduction reduced to 1,533,238. These variants were initially used for Hardy Weinberg (HWE) and linkage imbalance analysis. HWE analysis showed that most genetic variation in all chromosomes comply with HWE expectations. Overall, as shown in Table S1, 99.56% of the variants were in Hardy-Weinberg equilibrium, but only 0.44% deviated from the equilibrium. Analysis of linkage disequilibrium using the specified threshold showed that 19,469,198 out of the 22,827,466 variants exhibited high binding imbalances, corresponding to approximately 85% of the total mutants. In subsequent analyses, normalization of the VCF increased the number of variants to 27,935,252, consisting of 21,878,061 SNPs and 5,502,684 indels, splitting all multi-allen sites. These variants were distributed across all chromosomes, with the majority (94.96%) classified as “known” variants and approximately 5.04% classified as “new” (Table 1). The proportion of new variants ranges from about 3.90% to 5.71%, and is relatively consistent across most chromosomes, but chromosome Y is newly classified into almost half of its alleles (46.34%). Mitochondrial DNA (CHRM) also shows a higher percentage of novel variants (7.81%) than nuclear chromosomes.
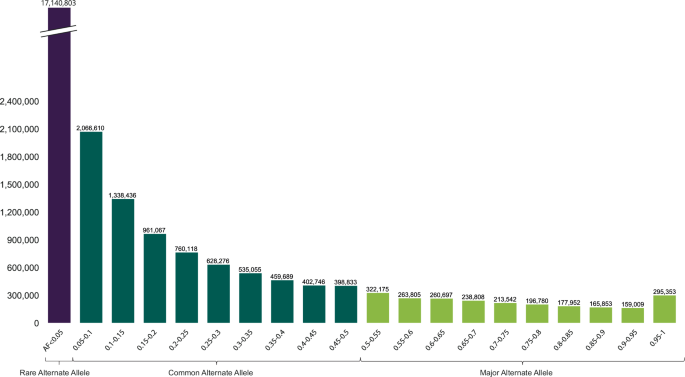
The 26,985,607 variants obtained after removing more than 11 genotypes were divided into three groups based on allele frequency (AF). The majority (61%) were rare alternative alleles (AF 1).

The histogram shows the count of variants per 5% AF interval. The most prominent peaks correspond to variants with rare alternative AFs below 5%, whereas rare or unobserved reference variants have 100% alternative AFs, indicating less common variants.
We analyzed variants using Clinvar Annotation to identify 231 pathogenic and possibly pathogenic changes, and identified the majority (205) as part of the exome. These changes include 167 single nucleotide variations (SNVs) and 64 insertions or deletions (indels) and affect 191 unique genes. Most of these variants are rare, with an allele frequency (MAF) of more than two-thirds. On average, each individual has 21 of these variants on 12-29. The mean allele frequency (AF) for these variants is 0.0598, with a range of 0.00458-0.9862.
The distribution of variants and their top-level results are shown in Figure 2 and Figure S1, respectively. These representations reveal different patterns of variant types across exomes. Chromosome 1 shows the highest total mutants (24,350), followed by chromosome 19 with 19,794 variants. In contrast, chromosome Y shows the lowest variant count (60 variants). SNVs dominate on most chromosomes, with their proportions ranging from 87.49% to 93.45% of variants. In particular, chromosome Y stands out compared to others with its percentage of insertion (13.33%) and complex variants (6.67%). Analysis of pathogenic mutants reveals significant concentrations for a particular chromosome. Chromosomes 1, 11, and 3 appear as hotspots with 28, 21, and 18 pathogenic variants, respectively, indicating potential clinically relevant regions. Chromosomes 6, 12, and 16 also show prominent pathogenic mutation counts ranging from 10 to 12 variants. Conversely, chromosomes 14, 15, and 21 show minimal pathogenic variants. Furthermore, we listed the most frequent variants (55 variants) that have a higher functional impact on Moroccan populations compared to gnomads (Supplementary Data 1).

From outer ring to inner ring: blue represents SNV distribution, green represents deletion, orange represents insertion, red depicts complex variants (scale adjusted for visibility). The innermost ring indicates pathogenic variants and their frequency in the Moroccan population.
Loss of functional analysis
Using VEP's Loftee Plugin17, 1086 variants with allelic frequencies (AFs) above 0.01 were predicted to cause high confidence loss of function (LOFs). These variations included 501 SNPs, 346 deletion, 210 insertions, and 29 complex variants. Narrows searches for common LOF variants in Moroccan samples (AF>0.05) and rare in other populations (GNOMAD Exome PRSS1, associated with hereditary pancreatitis.
Major Allele Reference Genomes in Morocco
The major allelic reference genome (MMARG) of the Moroccan population was based on 2,257,746 variants, including 1,907,253 SNPs and 350,493 indels. Compared to GRCH38, variant calls using MMARG showed consistently lower variant counts across all chromosomes (Table 2) (Fig. S2). The total number of variants detected using GRCH38 reference was 4,978,994, while the total count using MMARG was 2,737,930, with a difference of 2,241,064, which corresponds to a 45.01% rate. Chromosome Y showed the highest reduction of 64.57% followed by chromosome X at 52.78% and chromosome 21 at 51.87%. The lowest reduction was observed in chromosome M in 40.54%, chromosome 5 in 41.28% and 41.90% in chromosome 16.
Genetic relationships between Morocco and global populations
Genetic diversity in Moroccan populations was analyzed by comparing their genomic data with genomic data from the 1000 Genome Project and the Human Genome Diversity Project using a variety of statistical methods and analyses. Principal Component Analysis (PCA) places Moroccan populations and Mozabitans within the same cluster along the European African axis, showing strong genetic proximity between the two populations. Also, genetic proximity was observed in European and Middle Eastern clusters, and to a lesser extent American clusters, as shown in Figure 3A (see Supplementary Data 2 for more detailed visualization).

Principal Component Analysis (PCA) was conducted using data from 3,586 people representing various populations around the world. The points are color coded according to the super group. b Mixing results k = 19 show that Moroccan populations have zooms, indicating four main ancestral components. c. Heatmap of pairwise FST values between Moroccan genomes and various populations. The displayed value corresponds to FST multiplying by 1000. d. Boxplot of the total length of homozygosity (ROH) in Moroccans compared to other populations. The colour indicates a supergroup. The number of individuals per population is displayed in parentheses. The boxplot shows the median and lower/upper quartiles. A whisker represents the most extreme data points that do not exceed 1.5 times the interquartile range. Outliers are data points outside the whisker. Furthermore, P values comparing the average total length of ROH have been estimated using GGPUBR57. The selection of populations to calculate FST and ROHS was based on proximity to Moroccan populations based on PCA and mixing results. Results of PCA, FST, and ROH analyses were visualized using R58. Mixing results were visualized at Pong v 1.559.
PCA results were supported by mixed analysis (Figure S3). Since this value showed the lowest cross-validation (CV) error, we chose K = 19 to estimate the ancestors of the Moroccan population. It was found that 80% of the Moroccan variants analyzed consisted of four main ancestral components: North Africa (51.2%), Europe (10.9%), Middle East (10.7%), and West Africa (6.8%). Furthermore, these results show low genetic heterogeneity evidenced by minimal variation in the proportion of ancestral components between individuals (Fig. 3B).
Additionally, pairwise FST analyses were performed for genetic intimacy with Moroccan populations using a subset of populations from mixed analysis, including European, Africa, North Africa, and Middle Eastern populations. In total, 618 people from a population of 38 were included in the data set. This analysis revealed that Moroccans showed the lowest genetic distance to Mozabites (FST = 8.147), but the largest genetic distance was observed in the through population (FST = 139.996) (Fig. 3C) (Supplementary Data 3).
The mean total length of ROHS (>1 MB) in Moroccan populations (Supplementary Data 4) was comparable to that of Middle Eastern and Mozavite populations, with no significant differences observed (P≥0.05, Wilcoxon test) (Supplementary Data 5). These populations showed relatively large ROH compared to most other populations. This can be attributed to the widespread practice of kinship in these areas. Furthermore, the Luhija population had the shortest ROH, whereas the Kalitiana population showed the largest ROH (Fig. 3D).
Identification of mitochondria and Y DNA haplogroups
To further validate the findings of the previous findings, haplogroup analysis was performed using mitochondrial DNA (MT-DNA) and Y chromosomal markers. The mitochondrial haplogroup is Coudray et al. 23: European haplogroups (H, HV, R0, J, T, U, W), sub-Saharan African haploops (L0, L1, L2, L3), and North African genera (U6, M1). Our results show that of the 109 Moroccan samples analyzed, 73% showed European haploops (H (29.4%), U (15.6%), T (8.3%), and J (2.8%), not a recent historical event, but rather an Iberian Peninsula 24. Furthermore, 19% of the samples were sub-Saharan Africans, including L2 (27.3%), L3 (11%), and L1 (10.1%), while 8% of the mitochondrial haplogroups were attributed to the indigenous North African line M (5.5%) (Fig. 4A). Y chromosome analysis identified the E1B1B1(M35) haplogroup as more frequent in Moroccans. This lineage is also found at various frequencies in 25,26 North and East Africa (Fig. 5).

Total haplogroup frequency in the Morocco population. B DNA D-Loop Haplotype Network: A median coupled network comparing 109 Moroccans with populations in Africa, Europe and America. Green Circle shows the Moroccan haplogroup.

The bar chart shows the frequency of Y-chromosomal haplogroups found in a sample of 109 Moroccans. E1B1B1 is the most common (36.6%) followed by F (19.5%) and G2 (17.1%). Less frequent haplogroups include E1B1, R1, E1, R1B1, and K. Each haplogroup is represented and the corresponding proportions are shown.
Haplotype Network
In the first test, two prominent clusters emerged, effectively describing haplotypes in Africa and Europe. American haplotypes are primarily located within European clusters, but formed identifiable subclusters, particularly on the right side of the network, indicating representations within African clusters. In particular, Moroccan haplotypes are primarily consistent with European clusters, accounting for about 66% of the total Moroccan sample, with the remaining 34% being distributed in African haplotype clusters. Furthermore, Moroccan haplotypes demonstrated the formation of subclusters, accounting for approximately 24% of the total Moroccan sample, indicating within-group diversity (Fig. 4B).